COMPARATIVE ANALYSIS OF S494 MUTANTS
The side chain of S494 in wild-type 2019-nCoV-S1 (as shown in Figure 8A) is shorter and could not form direct contacts with any amino acid on ACE2. Therefore, mutations could make unlimited influence on the local region, and the RMSD values fluctuated greatly compared with the other systems.
(a) When S494 were mutated to reisdues ACFGILMNPQTVWYH, the sidechains from mutants were insufficient to form novel interactions with the amino acids in ACE2 (Figure 8A).
(b) For S494D and S494E, the mutated residue E494 and D494 could form hydrogen bonds with Q493, which weakened the charge distribution of K31-Q493-E35 in wild proteins, and this resulted in the decrease of binding modes between 2020-nCoV-S1 and ACE2 (Figure 8B).
(c) When S494 was mutated to K494, the interactions among Q493, K31 and E35 were not greatly affected. However, the new salt-bridges were observed between K494 and E35 which could potentially enhance the binding capacity of the 2019-nCoV-S1-ACE2 complex (Figure 8C).
(d) S394R completely disrupted the original endogenous interaction within the wild-type protein, resulting in the appearence of extremely strong salt-bridges: R394-E35 and E484-K31. All these results enchanced the interactions between 2019-nCoV-S1 and ACE2, which should be explored in details (Figure 8D).
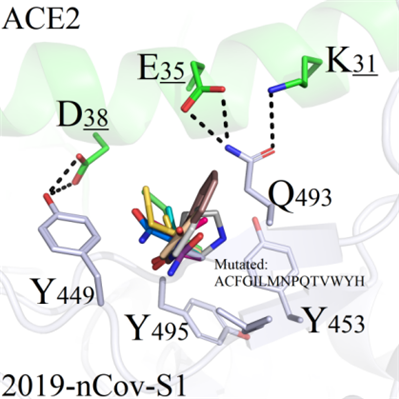 A
A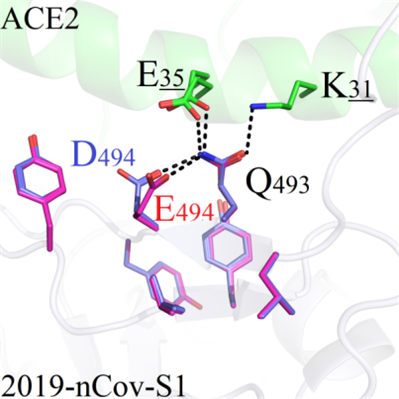 B
B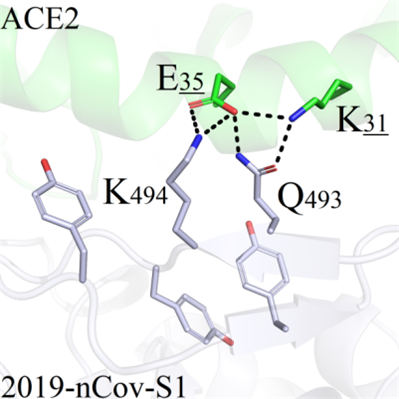 C
C D
DFigure 8. Comparison analysis of the complex structures between wild-type and mutant systems.