COMPARATIVE ANALYSIS OF Q493 MUTANTS
The amino acid Q493 of 2019-nCoV-S1 could form directly hydrogen bonds with residues E35 and K31 on ACE2 protein (Figure 6A). Therefore, mutations could trigger local environmental changes and significant fluctuations of RMSD values which were greater than 0.6.
The side chains of the amino acids ACGILNPV after mutations are shorter; therefore, they could not make up for the polarity effects existing in wild system (Figure 6A). The hydrophobic amino acids FMWY, with long side chains, were conflic with the hydrophilic residues E35 and K31, and the original interaction modes disappeared (Figure 6B). Mutations of Q493T and Q493S, both the hydroxyl group -OH on the two amino acids could form hydrogen bonds with E35 and K31 (Figure 6C). The amino acids D493/E493 could pull residue K31 from ACE2 closer to form new salt-bridging interactions (Figure 6D), and the same situations were also found in mutants Q493K, Q493H and Q493R (Figure 6E/F).
However, in terms of strong salt-bridging effects, the mutants Q493R, Q493H, Q493K, Q493E and Q493D were considered for subsequent binding energy analysis.
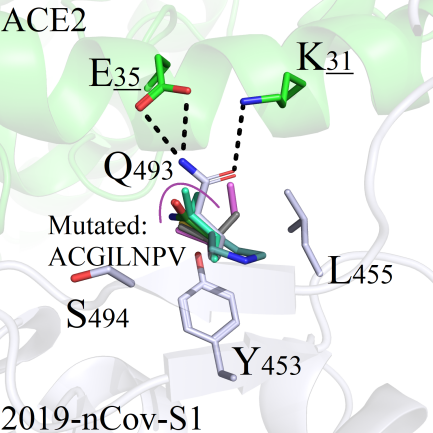 A
A B
B C
C D
D E
E F
FFigure 6. Comparison of protein structures between wild-type and mutant systems.