The mission of Drugs&Diseases Calculation is to provide the scientific community with a comprehensive,high-quality and freely accessible tool of drug-drug similarity calculation or disease-disease similarity calculation based on some drug features or disease features.
Dataset is built based on multiple open source online databases,such as Drugbank,SIDER,ChEMBL,PROMISUOUS.
Drugs A substance used to prevent, treat, and diagnose a disease.
Drugs data extracted from open-source database such as DrugBank,selected and analysed.
Diseases The self-stabilizing disorder regulates the process of abnormal life activities, and triggers a series of changes in metabolism, function and structure.
Diverse data sources.
 Drug-Drug Similarity
Drug-Drug Similarity
-
 ATC Code
ATC Code
The Anatomical Therapeutic Chemical (ATC) Classification System is used for the classification of active ingredients of drugs according to the organ or system on which they act and their therapeutic, pharmacological and chemical properties. It is controlled by the World Health Organization Collaborating Centre for Drug Statistics Methodology (WHOCC), and was first published in 1976.ATC code can be obtained from DrugBank database.
-
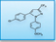 Chemical Structure
Chemical Structure
The chemical structure is a category that reflects the order of the atoms of each element inside the material molecule, that is, the way in which the atoms are connected and the order, and is the basis for understanding and mastering the chemical properties of the substance and the laws of chemical reaction. The chemical structure of the drug affects the molecular activity of the drug
-
 Side Effects
Side Effects
Drug side effects refers to pharmacological effects other than the therapeutic purpose that occurs after a therapeutic amount of a drug is applied.The database Sider saves side effect information
-
 Drug-Targets
Drug-Targets
Drug targets refer to biological macromolecules, such as some proteins and nucleic acids, which have pharmacodynamic functions in vivo and can be acted by drugs. Genes that encode target proteins are also known as target genes. Pre-determination of target molecules related to specific diseases is the basis of modern drug development.
Targets contain sequence, protein-protein interaction, gene ontology and so on.
 Drug-Drug Interaction
Drug-Drug Interaction
Drug-Drug interaction refers to the combined effects of two or more drugs taken by patients at the same time or in a certain period of time, which can enhance the efficacy or alleviate the side effects, weaken the efficacy or produce undesirable side effects. Enhanced effects include increased efficacy and increased toxicity, while weakened effects include reduced efficacy and toxicity. Therefore, in the clinical use of combined drugs, we should pay attention to the use of the characteristics of various drugs, give full play to the pharmacological role of each drug in the combined drugs, in order to achieve the best curative effect and minimize adverse drug reactions, so as to improve the safety of drug use.
 Disease-Disease Similarity
Disease-Disease Similarity
-
 Disease-gene association
Disease-gene association
Disease prevention can be carried out through the relationship between diseases and genes, and genetic information can reflect disease information to some extent. The similarity between diseases can be calculated by the relationship between disease-genes and gene ontology information.
-
 Disease ontology
Disease ontology
Disease Ontology (DO) is the official ontology of human disease.The Ontology Project is hosted by the Institute of Genomics at the University of Maryland School of Medicine.
Originally developed at Northwestern University in 2003, the Ontology Project aims to meet the needs of specific building ontology that covers all the disease concepts annotated in the biomedical resource pool within an ontological framework that can be extended to meet community needs.
Disease ontology is an OBO (Open Biomedical Ontology) foundry ontology.
The disease ontology identifier (DOID) consists of the prefix DOID: followed by a number, for example, Alzheimer's disease has a stable identifier DOID: 10652 -
 Phenotype
Phenotype
The phenotype can be any observable characteristic or trait of the disease, such as morphological, developmental, biochemical or physiological properties or behavior, without any suggestion of a mechanism. The clinical phenotype will be the manifestation of the disease in a given individual